 |
 |
AbstractBackground and ObjectivesWe evaluated differences in the pathophysiology of atopic and non-atopic eosinophilic nasal polyps and investigated their distinct inflammatory profiles.
Subjects and MethodA total of 36 patients were recruited: 10 controls (Group C), 14 with chronic rhinosinusitis with eosinophilic nasal polyps with atopy (Group A), and 12 without atopy (Group NA).
ResultsSerum eosinophil counts, total immunoglobulin E, eosinophil cationic protein levels, and tissue eosinophil counts were elevated in Groups A and NA vs. Group C. Real-time polymerase chain reaction results showed increased GATA-3, interleukin (IL)-4, IL-33 levels, but decreased levels of retinoic acid-related orphan receptor gamma t, IL-17 in Groups A and NA. Related to the regulatory T (T-reg) cell response, forkhead box P3 (Foxp3+) (A: p<0.001, NA: p<0.001) and IL-10 (A: p<0.001, NA: p<0.001) levels were elevated and transforming growth factor-╬▓ levels (A: p<0.001, NA: p<0.001) were decreased in Group A and Group NA in comparison to those in Group C. The Foxp3+ (p=0.001) and IL-10 (p<0.001) were significantly higher in Group A than in Group NA.
IntroductionNasal polyposis (NP) is a chronic inflammatory condition of the nasal mucosa with heterogeneous findings. The pathogenesis of NP is yet to be thoroughly revealed, it is presumed to be related to abundance of inflammatory cells. Eosinophilic inflammation is found in 65-90% of NP cases in Caucasians [1,2]. However, non-eosinophilic NP has been reported in more than half of NP patients in Korea, similar to the situation in neighboring Asian countries [3]. It has been reported that the NP in Western population demonstrates different immunopathological features from those in Asian countries [3-5].
Recent environmental and lifestyle changes of Asian population towards those of Western countries, led to increased prevalence of allergic diseases in Asia [6]. Consequently, prevalence of NP with eosinophilic and atopic NP has been on a continuous increase also in Korea [7]. A recent paper reported that 62.6% of NP in Korea was of the eosinophilic type [7].
However, the importance of the atopic influence in eosinophilic NP remains controversial. Therefore, we investigated the atopic characteristics and immunologic profiles of eosinophilic and non-eosinophilic NP using nasal mucosa and peripheral blood tests in this study. This endotype analysis might be helpful in understanding the development and response of biotherapeutics.
Subjects and MethodsPatientsThe collection of human tissue samples used was approved by the Institutional Review Board of Seoul St. MaryŌĆÖs Hospital (KC12TISI0523). All patients provided written informed consent prior to the study.
The atopic status of the patients was evaluated using a skinprick test and/or a multiple allergen simultaneous test (CLA Allergen-specific immunoglobulin E (IgE) assay, Hitachi Chemical Diagnostics, Mountain View, CA, USA) to a panel of aeroallergens (e.g., pollen, dust mites, pets, molds, and cockroaches). We used wheal average diameter of 3 mm or +++ of skin-prick test [8,9] and class Ōēź1 of multiple allergen simultaneous test [10] for the cut-off to divided atopic test results. Chronic rhinosinusitis with NP (CRSwNP) was diagnosed based on history taking, clinical examination, nasal endoscopy, and a CT scan according to European Position Paper on Rhinosinusitis and Nasal Polyps guideline [1]. Exclusion criteria included immunodeficiency, cystic fibrosis, bronchiectasis, chronic obstructive pulmonary disease, diabetes mellitus, neoplasia, oral steroid treatment, upper airway infection within 1 month, and acute exacerbation of asthma.
Preoperative paranasal sinus (PNS) CT was performed for all patients. PNS CT was performed in the axial projection, obtaining serial axial images (0.6 mm thick). Lund-Mackay scores were checked using PNS CT preoperatively.
Inferior turbinate tissue collected from patients undergoing septoplasty or septorhinoplasty were used as controls (Group C; n=10) [11-14]. During functional endoscopic sinus surgery procedures samples of nasal polyps (14 atopy patients with eosinophilic NP, Group A, and 12 non-atopy patients with eosinophilic NP, Group NA) were obtained.
Nasal tissue evaluationSamples of nasal polyp were fixed in 4% paraformaldehyde at 4┬░C for 3 days, dehydrated by passing through gradient of alcohol, and embedded in paraffin wax blocks. Paraffin blocks were cut in into 4-╬╝m sections and stained with hematoxylin and eosin for evaluation of morphology and number of eosinophils in the lamina propria. Eosinophils count was done under light microscope at ├Ś400 magnification (Olympus CX-40 microscope; Olympus, Tokyo, Japan). Two independent observers blinded to the diagnosis and clinical data analyzed four random fields in each section. Mean value of eosinophil count in four fields of each sample was used as the final score.
NPs with eosinophil count exceeding 10% of the total infiltrating cell count per high-power field, were classified as eosinophilic [15]. Three different processing was done on tissue samples; snap-freezing in liquid nitrogen at -80┬░C for ribonucleic acid (RNA) isolation, isolating protein from tissue, and overnight fixing in freshly prepared fixative of 4% paraformaldehyde in phosphate buffered saline (pH 7.4) followed by embedding in paraffin wax for histological staining.
Eosinophil cationic protein and total immunoglobulin E levels in serum and eosinophil counts in peripheral bloodEnzyme-linked immunosorbent assay (ELISA) kit (Indoor Biotechnologies, Manchester, UK) was used for measurement of levels of total IgE and eosinophilic cationic protein (ECP). The minimum detectable dose (sensitivity) of human ECP was 0.39 ng/mL. The sensitivity of the ELISA, or lower limit of detection, was defined as the lowest protein concentration that could be differentiated from zero. Sensitivity was presented as mean optical density value of 20 replicates of zero plus three standard deviations.
ELISA is a quantitative sandwich enzyme immunoassay. Samples are pipetted into wells of microplate pre-coated with specific antibody for ECP, so that ECP is bound to the immobilized antibody. After removing other unbound substances, biotin-conjugated antibody specific for ECP is applied. Then avidin- conjugated horseradish peroxidase is added, and after washing unbound avidin-enzyme reagent, color of the sample changes in proportion to the concentration of ECP bound to the antibody. Eosinophil counts were measured by ADVIA 120 Hematology System (Bayer, Tarrytown, NY, USA).
Real-time polymerase chain reactionThe nasal mucosa was removed and prepared for real-time PCR detection. T-bet and interferon gamma (IFN-╬│) for the T helper (Th) 1 immune reaction, GATA-3 and interleukin (IL)-4, IL-33 for the Th2 immune reaction, retinoic acid-related orphan receptor gamma t (ROR-╬│t) and IL-17 for the Th17 immune reaction, Foxp3, IL-10 and transforming growth factor (TGF)-╬▓ for the T-reg immune reaction, were measured by real-time PCR. RNA extraction was done by TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and reverse-transcribed using random primers (TaKaRa, Otsu, Japan). Following oligonucleotide primer sequences were used: IFN-╬│ forward primer, 5ŌĆÖTCCCATGGGTTGTGTGTTTA-3ŌĆÖ, IFN-╬│ reverse primer, 5ŌĆÖAAGCACCAGGCATGAAATCT-3ŌĆÖ, T-bet3 forward primer, 5ŌĆÖTCAGCATGAAGCCTGCATTCTTGC-3ŌĆÖ, T-bet reverse primer, 5ŌĆÖACAGCTATGAGGCTGAGTTTC GA-3ŌĆÖ, GATA-3 forward primer, 5ŌĆÖCAAAATGAACGGA CAGAACCG-3ŌĆÖ, GATA-3 reverse primer, 5ŌĆÖGCTCTCCTG GCTGCAGACA-3ŌĆÖ, IL-4 forward primer, 5ŌĆÖCGAGTTGAC CGTAACAGACAT-3ŌĆÖ, IL-4 reverse primer, 5ŌĆÖCGTCTT TAGCCTTTCCAAGAAG-3ŌĆÖ, IL-17 forward primer, 5ŌĆÖAC CAATCCCAAAAGGTCCTC-3ŌĆÖ, IL-17 reverse primer, 5ŌĆÖGGGGACAGAGTTCATGTGGT-3ŌĆÖ, ROR-╬│t forward primer; 5ŌĆÖGCTGTGATCTTGCCCAGAAC -3ŌĆÖ, ROR-╬│t reverse primer, 5ŌĆÖCTGCCCATCATTGCTGTTAATCC-3ŌĆÖ, IL10 forward primer, 5ŌĆÖGATGCCTTCAGCAGAGTGAA-3ŌĆÖ, IL-10 reverse primer, 5ŌĆÖ GCAACCCAGGTAACCCTTA AA-3ŌĆÖ, TGF-╬▓1 forward primer, 5ŌĆÖCGAGCCTGAGGCC GACTAC-3ŌĆÖ, TGF-╬▓1 reverse primer, 5ŌĆÖAGATTTCGTTGT GGGTTTCCA-3ŌĆÖ, Foxp3 forward primer, 5ŌĆÖGAGAAGCT GAGTGCCATGCA-3ŌĆÖ, Foxp3 reverse primer, 5ŌĆÖAGGAGCC CTTGTCGGATGAT-3ŌĆÖ, IL-33 forward primer, 5ŌĆÖCAAAGA AGTTTGCCCCATGT-3ŌĆÖ, IL-33 reverse primer, 5ŌĆÖAAGG CAAAGCACTCCACAGT-3ŌĆÖ, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward primer, 5ŌĆÖ-GTGGATA TTGTTGCCATCAATGACC-3ŌĆÖ, and GAPDH reverse primer, 5ŌĆÖ-GCCCCAGCCTTCTTCATGGTG GT-3ŌĆÖ.
Results were normalized by comparison to expression rate of GAPDH, and represented as fold increase over the control group.
Western blot analysisT-bet, GATA-3, ROR-╬│t, IL-10, and Foxp3 expression of samples were detected by Western blot analysis. The antibodies used were T-bet antibody sc-21003 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); GATA-3 antibody ab106625; IL-10 antibody ab34843; Foxp3 antibody 14-6988; and ROR╬│t antibody 14-6988 (eBioscience, San Diego, CA, USA). Results were presented as grayscale ratio of target gene compared to GAPDH.
Statistical analysisParameters are presented as means┬▒standard deviation. Differences between groups were evaluated by the KruskalWallis test. Analysis of statistical significance was done by comparing ranked parameters with one-way analysis of variance and BonferroniŌĆÖs multiple comparison tests (PASW Statistics 24; SPSS Inc., Chicago, IL, USA). In all analyses, p<0.05 was considered statistically significant.
ResultsDemographic data and Lund-Mackay scoresThe mean age and male/female ratios of the C, A, and NA groups were 59.5┬▒17.2 (8/2), 45.5┬▒11.8 (10/2), and 43.22┬▒17.65 (7/7), respectively. The mean Lund-Mackay scores of the three groups were 0, 13.83┬▒6.26, and 10.64┬▒7.68, respectively (p=0.288). The Lund-Mackay score differences between the A and NA groups were not significant. There were three patients with atopy in the Group C.
Blood eosinophil counts, total IgE, and ECPGroup A showed significantly higher total IgE level (34.64┬▒41.23 ng/mL, p<0.001) and NA (20.04┬▒27.37 ng/mL, p<0.001) (Fig. 1A). ECP levels were significantly higher in Group A (58.47┬▒36.92 ng/mL) than in Groups C (19.47┬▒17.37 ng/mL, p=0.003) or NA (28.91┬▒33.21 ng/mL, p=0.037) (Fig. 1B). Blood eosinophil counts were significantly elevated in Group A (50.53┬▒28.3├Ś107/mL) than in Groups C (11.83┬▒7.80├Ś107/mL, p=0.009) or NA (15.33┬▒11.93├Ś107/mL, p=0.005) (Fig. 1C).
Real-time PCR and Western blottingRegarding Th1 response, IFN-╬│ messenger ribonucleic acid (mRNA) levels were not significantly different among the groups (C vs. A, p=1.000; C vs. NA, p=1.000) (Fig. 3A) and T-bet mRNA expression was significantly higher in Group C than in Groups A (p=0.003) or NA (p=0.001) (Fig. 3B). Regarding Th2 response, GATA-3 mRNA expression was significantly higher in Groups A (p<0.001) and NA (p<0.001) than in Group C (Fig. 3C), IL-4 mRNA expression was significantly higher in Groups A (p<0.001) and NA (p<0.001) than in Group C (Fig. 3D), and IL-33 mRNA expression was significantly higher in Groups A (p<0.001) and NA (p<0.001) than in Group C (Fig. 3E). Regarding Th17 response, IL-17 mRNA expression was significantly lower in Groups A (p<0.001) and NA (p<0.001) than in Group C (Fig. 3F), and ROR-╬│t mRNA expression was significantly lower in Groups A (p<0.001) and NA (p<0.001) than in Group C (Fig. 3G). Regarding regulatory immune response, TGF-╬▓ mRNA expression was significantly lower in Groups NA (p<0.001) and A (p<0.001) than in Group C (Fig. 3H), IL-10 mRNA expression was significantly higher in Group A than in Groups C (p<0.001) or NA (p<0.001) (Fig. 3I), and Foxp3 mRNA expression levels were significantly higher in Group A than in Groups C (p<0.001) or NA (p=0.001) (Fig. 3J).
Western blotting was conducted to confirm the expression patterns of transcription factors and cytokines mentioned above. A quantitative test was not performed, but the overall patterns of each factor in grayscale were analyzed (Fig. 4).
DiscussionThe eosinophilic CRSwNP phenotype is related to extensive disease and a poor prognosis compared with other phenotypes [16]. Our results was consistent with previous results: eosinophilic nasal polyp patients showed eosinophilic inflammation (elevated IL-4, ECP, and total IgE), and significantly increased GATA-3 mRNA levels in the NP. However, current treatment strategies with anti-inflammatory and/or antibiotic therapy or surgical removal of the lesion to improve sinus drainage are less effective in eosinophilic CRSwNP patients. This lack of treatment efficacy highlights the need to find the endotype for further fundamental and clinical research on eosinophilic NP. The role of the immune response in the pathogenesis of mucosal remodeling, a key problem in eosinophilic NP, is of particular current interest.
Some studies have presented relationships between atopy and eosinophilic inflammation [17,18]. Also, it has been reported that the prognosis is worse in CRSwNP, especially in the presence of atopy [19]. Additionally, there is a report that the atopy factor is related to relapse after functional endoscopic sinus surgery [20]. In our study, atopic eosinophilic NP showed more enhanced eosinophilic inflammation, such as ECP and total IgE, than non-atopic eosinophilic NP. However, there was no difference in the expression of Th1, Th2, and Th17 markers, and Lund-Mackay scores were not significantly different between Groups A and NA. An interesting finding was the Foxp3 mRNA results. CD4+ CD25+ Foxp3+ cells are accepted as unique to the regulatory T (T-reg) cell lineage. Foxp3 is not only essential in T-reg development, but also performs a suppressive function in controlling the transcription of proinflammatory cytokines, such as Th2 and Th17-related cytokines [21,22]. However, in a situation in which an allergic reaction is actively occurring, Th2 markers may be high even if the T-reg response is increased as a defense response for controlling the Th2 reponse. Thus, induction of Foxp3+ T-regs and downregulation of Th2 and Th17 markers in patients with NP are indicative of an active immune-regulatory response at the site of persistent mucosal inflammation [23]. This may explain why no alteration in the systemic Th1 and Th2 cell populations was observed between Groups A and NA. TGF-╬▓ is known to play a role in mucosal remodeling, modulating extracellular matrix production and the activity of degrading proteases, and shows decreased expression in CRSwNP [24]. This discrepancy might have originated from IL-10 being induced by the chronic mucosal inflammatory condition [25].
In summary, we analyzed differences in the pathophysiology of atopic and non-atopic eosinophilic NP and their distinct inflammatory profiles. Increased T-reg cytokine production in the atopic group suggested that T-regs are stimulated by the allergic reaction. In both the atopic and non-atopic eosinophilic NP groups, the levels of cytokines secreted by Th2 cells and eosinophils were increased significantly, whereas the levels of Th17 cytokines decreased. These results suggest that the immunological activities of Th2 cells, eosinophils, and Th17 cells affect eosinophilic NP development. It will be important to further investigate the role of T-reg induction in atopic eosinophilic NP patients. However, since these results were conducted on patients visiting tertiary hospitals, attention should be paid to interpretation as selection biases may occur. Also, the immunologic results need to be carefully interpreted, because various factors including demographics and bias could affect these results.
T-reg cells and IL-10 may be major factors differentiating the pathophysiology of atopic and non-atopic eosinophilic nasal polyps. Moreover, the Th2 and Th17 profiles might be important in the development of eosinophilic nasal polyps.
NotesAuthor Contribution Conceptualization: Do Hyun Kim, Soo Whan Kim. Data curation: Boo-Young Kim, Il Hwan Lee, Sung Won Kim. Formal analysis: Boo-Young Kim, Do Hyun Kim. Methodology: Boo-Young Kim, Do Hyun Kim. Project administration: Soo Whan Kim. Resources: Soo Whan Kim. Supervision: Sung Won Kim, Soo Whan Kim. Validation: Sung Won Kim, Soo Whan Kim. WritingŌĆöoriginal draft: Boo-Young Kim, Do Hyun Kim. WritingŌĆöreview & editing: all authors. Fig.┬Ā1.Total IgE (A), ECP (B), and eosinophil counts (C) in the serum of each group. *C vs. A, ŌĆĀA vs. NA. C: controls, A: atopy patients with eosinophilic nasal polyp, NA: non-atopy patients with eosinophilic nasal polyp. IgE: immunoglobulin E, ECP: eosinophil cationic protein. 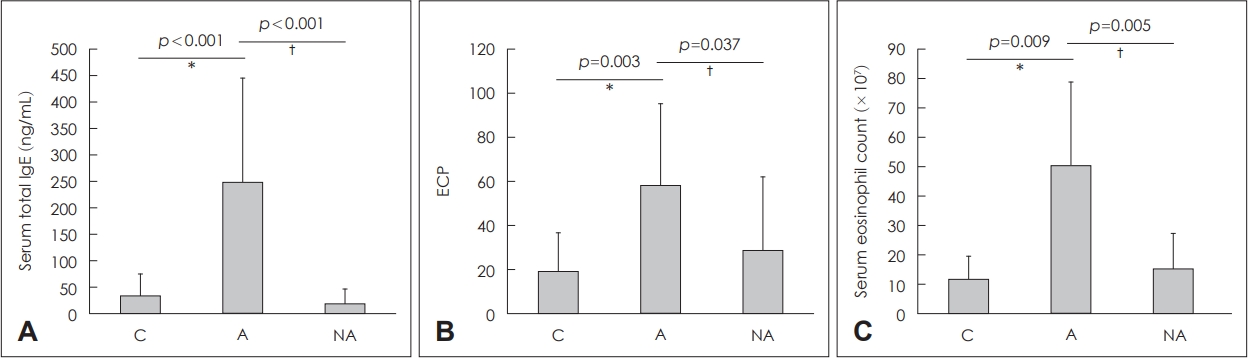
Fig.┬Ā2.Infiltration of eosinophils and eosinophil counts in the nasal mucosa. Infiltration of eosinophils (arrow) in each group: control (A), atopy patients with eosinophilic nasal polyps (B), non-atopy patients with eosinophilic nasal polyps (C) (hematoxylin and eosin staining, original magnification, ├Ś400). Eosinophil counts of each study group (D). *C vs. A, ŌĆĀC vs. NA. C: controls, A: atopy patients with eosinophilic nasal polyp, NA: non-atopy patients with eosinophilic nasal polyp. 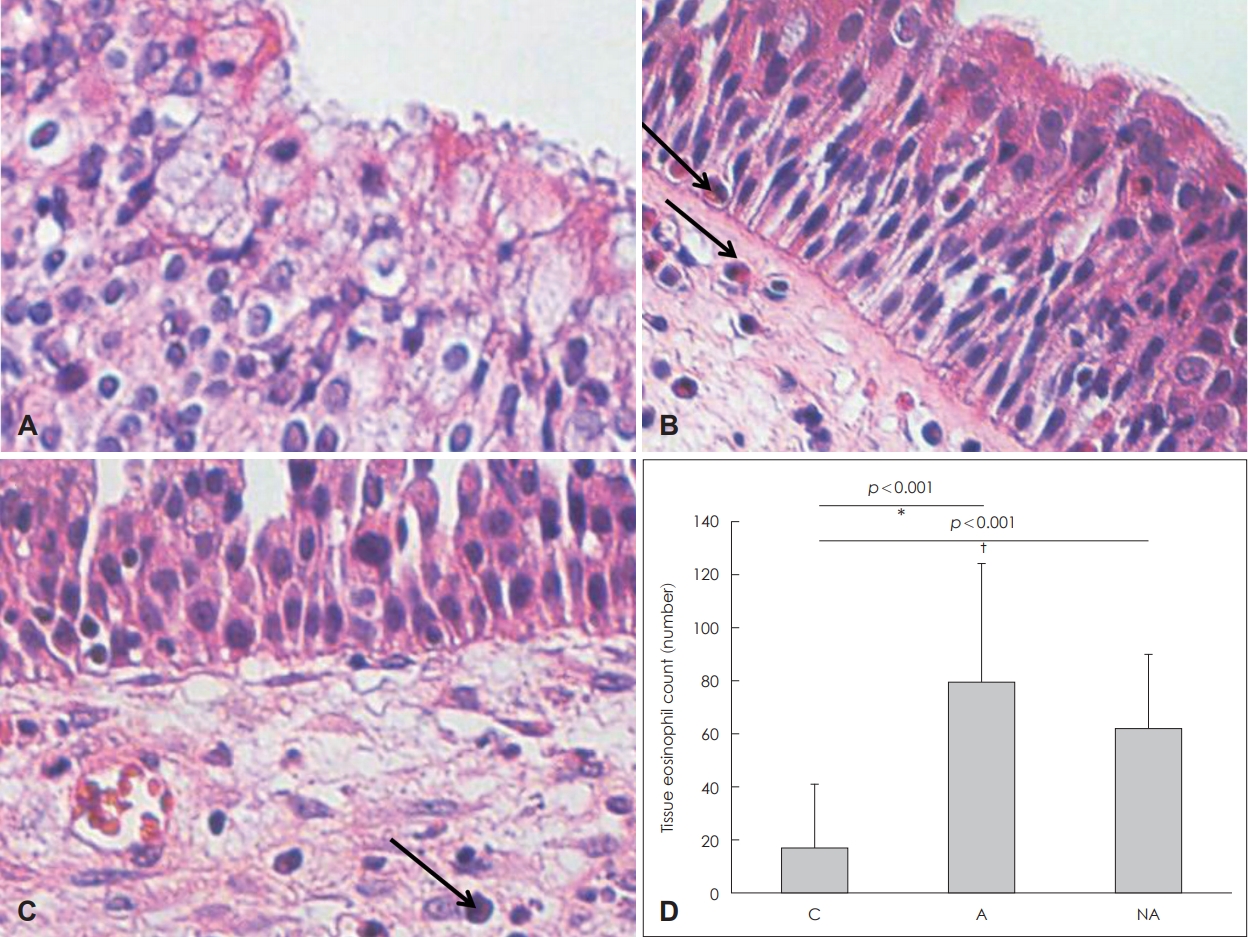
Fig.┬Ā3.Quantitative analysis by real-time PCR of cytokine transcript levels in the nasal mucosa of each group. IFN-╬│ (A), T-bet (B), GATA-3 for the T helper (Th) 1 immune reaction, (C), IL-4 (D), IL-33 (E) for the Th2 immune reaction, IL-17 (F), ROR-╬│t (G) for the Th17 immune reaction, TGF-╬▓ (H), IL-10 (I), and Foxp3 (J) for the T-reg immune reaction. *C vs. A, ŌĆĀC vs. NA, ŌĆĪA vs. NA. C: control, A: atopy patients with eosinophilic nasal polyps, NA: non-atopy patients with eosinophilic nasal polyps. IFN-╬│: interferon gamma, mRNA: messenger ribonucleic acid, IL: interleukin, ROR-╬│t: retinoic acid-related orphan receptor gamma t, TGF-╬▓: transforming growth factor-╬▓, Foxp3: forkhead box P3. 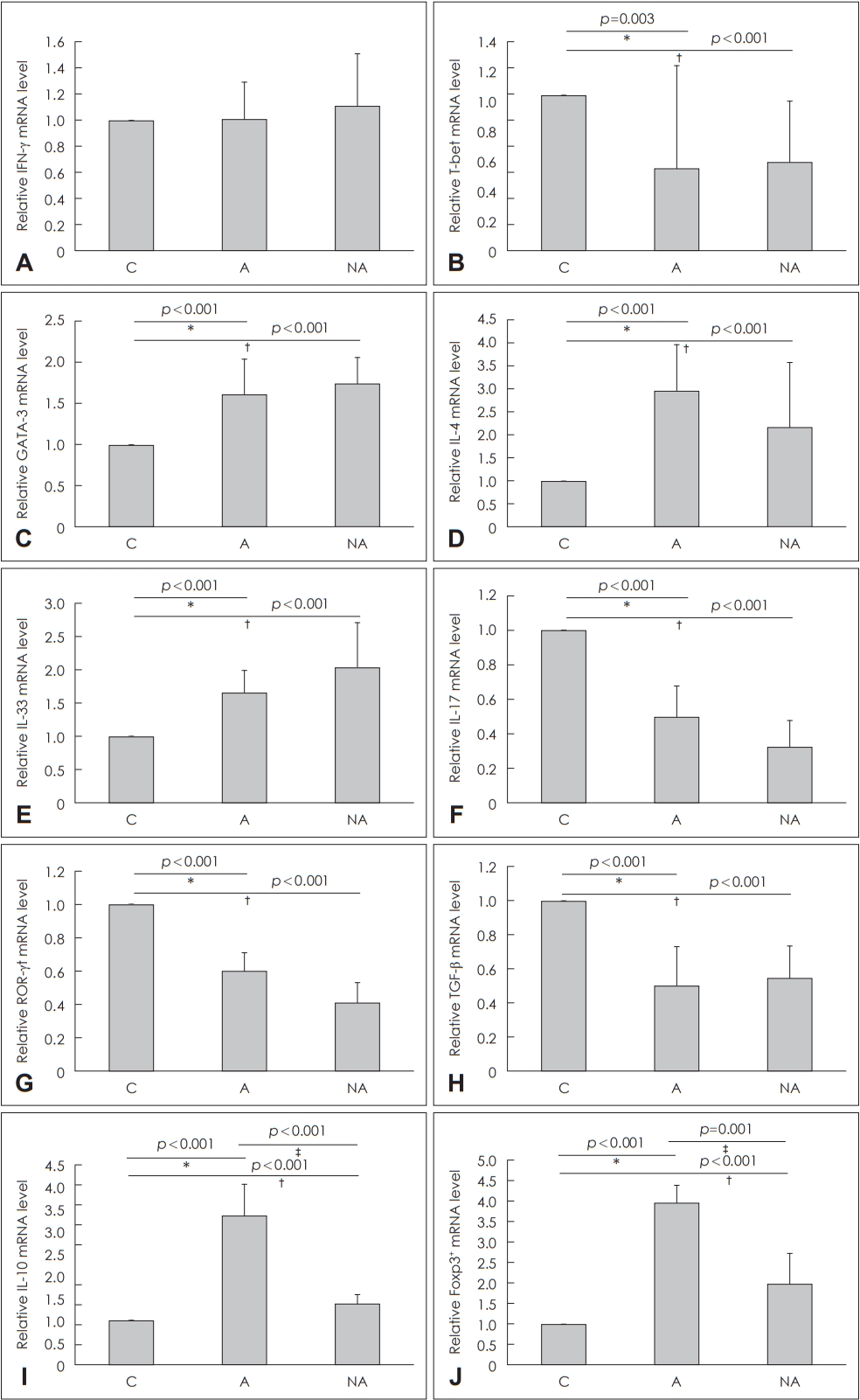
Fig.┬Ā4.Cytokines and transcription factors measured by Western blotting to confirm protein levels. C: control, A: atopy patients with eosinophilic nasal polyps, NA: non-atopy patients with eosinophilic nasal polyps. Foxp3: forkhead box P3, IL: interleukin, ROR-╬│t: retinoic acid-related orphan receptor gamma t, GAPDH: glyceraldehyde 3-phosphate dehydrogenase. 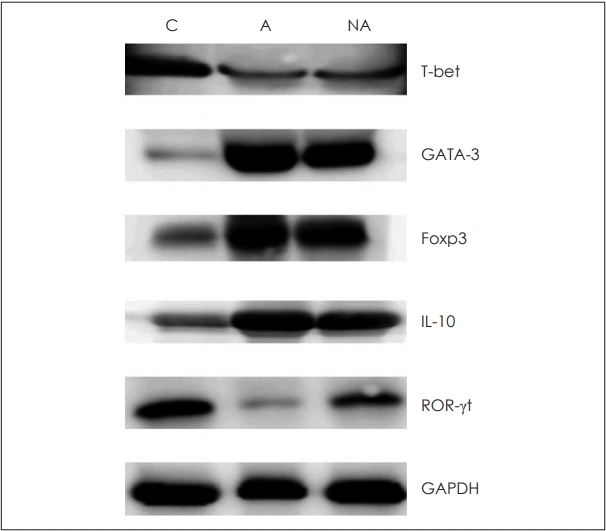
REFERENCES1. Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol 2012;50 Suppl 23.
2. Meltzer EO, Hamilos DL, Hadley JA, Lanza DC, Marple BF, Nicklas RA, et al. Rhinosinusitis: Establishing definitions for clinical research and patient care. J Allergy Clin Immunol 2004;114(6 Suppl):155-212.
3. Kim JW, Hong SL, Kim YK, Lee CH, Min YG, Rhee CS. Histological and immunological features of non-eosinophilic nasal polyps. Otolaryngol Head Neck Surg 2007;137(6):925-30.
4. Soler ZM, Mace JC, Litvack JR, Smith TL. Chronic rhinosinusitis, race, and ethnicity. Am J Rhinol Allergy 2012;26(2):110-6.
5. Zhang N, Van Zele T, Perez-Novo C, Van Bruaene N, Holtappels G, DeRuyck N, et al. Different types of T-effector cells orchestrate mucosal inflammation in chronic sinus disease. J Allergy Clin Immunol 2008;122(5):961-8.
6. Park HS, Choi GS, Cho JS, Kim YY. Epidemiology and current status of allergic rhinitis, asthma, and associated allergic diseases in Korea: ARIA Asia-Pacific workshop report. Asian Pac J Allergy Immunol 2009;27(2-3):167-71.
7. Shin SH, Ye MK, Kim JK, Cho CH. Histological characteristics of chronic rhinosinusitis with nasal polyps: Recent 10-year experience of a single center in Daegu, Korea. Am J Rhinol Allergy 2014;28(2):95-8.
8. van der Valk JP, Gerth van Wijk R, Hoorn E, Groenendijk L, Groenendijk IM, de Jong NW. Measurement and interpretation of skin prick test results. Clin Transl Allergy 2016;6:8.
9. Kim DY, Park SY, Hong SB, Lee OH, Choi GC, Cho JH. Comparison of multiple allergen simultaneous test and ImmunoCAP for the diagnosis of allergic rhinitis. Korean J Otorhinolaryngol-Head Neck Surg 2018;61(1):29-34.
10. Han M, Shin S, Park H, Park KU, Park MH, Song EY. Comparison of three multiple allergen simultaneous tests: RIDA allergy screen, MAST optigen, and polycheck allergy. Biomed Res Int 2013;2013:340513.
11. Lee JY, Lee SH, Lee HM, Lee SH, Jung HH, Lee SW, et al. Analysis of gene expression profiles of normal human nasal mucosa and nasal polyp tissues by SAGE. J Allergy Clin Immunol 2006;118(1):134-42.
12. Rhee CK, Jang YJ. Arachidonic acid metabolites in antrochoanal polyp and the nasal polyp associated with chronic paranasal sinusitis. Korean J Otolaryngol 1999;42(1):54-7.
13. Meng J, Zhou P, Liu Y, Liu F, Yi X, Liu S, et al. The development of nasal polyp disease involves early nasal mucosal inflammation and remodelling. PLoS One 2013;8(12):e82373.
14. Simon HU, Yousefi S, Schranz C, Schapowal A, Bachert C, Blaser K. Direct demonstration of delayed eosinophil apoptosis as a mechanism causing tissue eosinophilia. J Immunol 1997;158(8):3902-8.
15. Cao PP, Li HB, Wang BF, Wang SB, You XJ, Cui YH, et al. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol 2009;124(3):478-84, 484.e1-2.
16. Ikeda K, Shiozawa A, Ono N, Kusunoki T, Hirotsu M, Homma H, et al. Subclassification of chronic rhinosinusitis with nasal polyp based on eosinophil and neutrophil. Laryngoscope 2013;123(11):E1-9.
17. Ba L, Du J, Liu F, Yang F, Han M, Liu S, et al. Distinct inflammatory profiles in atopic and nonatopic patients with chronic rhinosinustis accompanied by nasal polyps in Western China. Allergy Asthma Immunol Res 2015;7(4):346-58.
18. Huang Z, Nayak JV, Sun Y, Huang Q, Zhou B. Peripheral blood T-helper cells and eosinophil populations in patients with atopic and nonatopic chronic rhinosinusitis. Am J Rhinol Allergy 2017;31(1):8-12.
19. Probst L, Stoney P, Jeney E, Hawke M. Nasal polyps, bronchial asthma and aspirin sensitivity. J Otolaryngol 1992;21(1):60-5.
20. Yacoub MR, Trimarchi M, Cremona G, Dal Farra S, Ramirez GA, Canti V, et al. Are atopy and eosinophilic bronchial inflammation associated with relapsing forms of chronic rhinosinusitis with nasal polyps? Clin Mol Allergy 2015;13(1):23.
21. Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: Differentiation, specification, subphenotypes. Nat Immunol 2009;10(7):689-95.
22. Sharma S, Watanabe S, Sivam A, Wang J, Neuwirth SJ, Perez RI, et al. Peripheral blood and tissue T regulatory cells in chronic rhinosinusitis. Am J Rhinol Allergy 2012;26(5):371-9.
23. Ling EM, Smith T, Nguyen XD, Pridgeon C, Dallman M, Arbery J, et al. Relation of CD4+CD25+ regulatory T-cell suppression of allergen-driven T-cell activation to atopic status and expression of allergic disease. Lancet 2004;363(9409):608-15.
|
|
||||||||||||||||||||||||||||||||||||||||||

 |
 |