 |
 |
AbstractRecently, real-time polymerase chain reaction (PCR) using the U-TOPвДҐHL Genotyping Kit has been introduced to detect genetic hearing loss caused by certain type of gene variants popularly found in Korea. The mitochondrial 12S ribosomal ribonucleic acid (rRNA) genes are related to aminoglycoside induced or non-syndromic, sensorineural hearing loss. Among them, 1555A>G is commonly found and reported worldwide. We are presenting the case of a mother and a son, who were screened by real-time PCR using the U-TOPвДҐHL Genotyping Kit and were found both to have the mitochondrial 12s rRNA 1555A>G variant with a different hearing loss phenotype. This report encourages clinicians to use this or similar screen methods for patients with familial hearing loss.
IntroductionGenetic sensorineural hearing loss is very common. It is responsible for 50% of all congenital hearing loss and for a small portion of progressive hearing loss, which could be of juvenile or adult onset. In cases of progressive hearing loss with no loss at birth, it is not easy to differentiate from environmental and age-related hearing loss. Due to low screening rates, testing every subject with sensorineural hearing loss is not cost effective.
Recently, real-time PCR using the U-TOPвДҐHL Genotyping Kit (SeaSun Biomaterials, Daejeon, Korea) has been developed to screen for genetic hearing loss among all sensorineural hearing loss subjects [1]. This kit screens certain types of genes that are common among Korean genetic hearing loss patients and is less expensive than other tests.
The mitochondrial 12S ribosomal RNA (rRNA) gene is related to mutations associated with aminoglycoside-induced or non-syndromic sensorineural hearing loss. Among them, 1555A>G is commonly found and reported worldwide [2-5]. Part of the effect of this mutation on the phenotype of the genetic condition is affected by use of specific aminoglycoside antibiotics. Limiting use of aminoglycosides in this group may prevent further hearing loss resulting from the variant.
In the present manuscript, we present the cases of a mother and son who were screened with real-time PCR using the U-TOPвДҐHL Genotyping Kit and found to have the mitochondrial 12s rRNA 1555A>G variant with different hearing loss phenotypes. This report encourages clinicians to use this or similar screening methods for patients with familial hearing loss.
CaseA 66-year-old female patient and her son (45 years old) without any past medical history visited the clinic. The mother visited due to progressive hearing loss, and the son did not have any auditory symptoms but was worried about progressive hearing loss in his future. There was no history of ototixic antibiotic treatment in either subject. Other symptoms such as dizziness and tinnitus were not obvious in either subject. Endoscopic exam of her tympanic membrane did not reveal any abnormalities. Their familial history of hearing loss was observed, with maternal genetic tendency (Fig. 1).
For initial functional evaluations, pure-tone audiology and speech audiometry were performed in both subjects. Aurical Plus (GN Otometrics, Denmark) was used, and frequencies from 0.5 to 8 kHz were assessed. Hearing thresholds of 0.5, 1, 2, and 4 kHz were averaged [6]. Profound hearing loss was observed in the mother; the right side hearing threshold was 92 dB HL with a descending type audiometric configuration (worse hearing), and the left side hearing threshold was 69 dB HL with a descending type audiometric configuration (better hearing). Word recognition scores, which show the percentage of correct response for single-syllable words at the most comfortable level of sound, were 44% (right side) and 72% (left side) (Fig. 2). In contrast to the profound hearing threshold shift in the mother, pure-tone audiometry thresholds were normal in both ears of her son.
For objective measurement of hearing, the auditory brainstem response, which is the averaged evoked potential due to repetitive sound stimuli, was performed. For auditory brainstem response, the Auditory evoked potential system navigation Pro, Version 7.0.0 (Bio-logic System Corp., Mundelein, IL, USA) was used with a rate of 13.3/s and a sound stimulus of alternating clicks broadcast through an inserted earphone (Bio-logic Insert Earphones 580-SINSER, Natus, San Carlos, CA, USA). Auditory brainstem response thresholds were no response (right side) and 70 dB nHL (left side), respectively, in the mother. The son did not undergo this test.
Next, a genetic hearing loss evaluation was performed. With peripheral blood, we tested DNA samples of the mother and the son collected at Dankook University Hospital. The U-TOPвДҐHL Genotyping Kit and conventional Sanger sequencing were used. Real-time PCR using the U-TOPвДҐHL Genotyping Kit and Sanger sequencing were performed as in a previous study [1]. Real-time PCR was performed with a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The sequences of primers and peptide nucleic acid (PNA) probes used in real-time PCR are shown in Table 1. The reaction conditions were 95¬∞C for 10 min followed by 42 cycles of 95¬∞C for 30 s, 58¬∞C for 45 s, and 72¬∞C for 45 s. Melting point analyses were followed. We performed a denaturation step of 95¬∞C for 5 min, 1 min hybridization steps at 75¬∞C, 55¬∞C, and 45¬∞C, and a 1¬∞C stepwise temperature increase from 20¬∞C to 85¬∞C, with a 5 s interval between each step. The data were analyzed with Bio-Rad CFX Manager v1.6 software (Bio-Rad). Variants were found based on the fluorescence signal of detection probes and corresponding melting temperatures (Tm).
For Sanger sequencing, PCR was performed in 20 ќЉL reactions containing 3 ќЉL purified gDNA (15 ng/ќЉL), 10 ќЉL 2√ЧqPCR PreMix (SeaSun Biomaterials), and 0.5 ќЉL each of a forward and reverse primer (10 mM). Thermocycling consisted of 40 cycles at 95¬∞C for 30 s, 58¬∞C for 45 s, 72¬∞C for 45 s, and 72¬∞C for 5 min. PCR products were sequenced using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA), according to the manufacturerвАЩs instructions. The sequencing primers were the same as the PCR primers (Table 1). Sequence data were analyzed using an ABI 3500XL DNA Analyzer (Applied Biosystems).
Both showed a 1555A>G variants with homoplasmy in the MTRNR1 gene (Fig. 3). The results of Sanger sequencing were the same as those of the U-TOPвДҐHL Genotyping Kit (Fig. 4).
After confirmation of the etiology and characteristics of hearing loss, for hearing rehabilitation, a hearing aid was recommended and fit in the left ear of the mother. Left hearing threshold was substantially decreased from 69 (unaided) to 48 (aided) dB HL, as shown by functional audiometry (Fig. 2). The word recognition score in the left ear at 55 dB HL was 52% without the hearing aid but improved to 76% with the hearing aid. We recommended the mother and son avoid use of aminoglycoside antibiotics.
DiscussionThese two cases show two different phenotypes of the same genetic condition. The mother showed progressive and profound hearing loss in both ears from her youth, but her son, who was old enough to have symptoms of hearing loss [7], showed no hearing loss despite the same genetic condition. This variable phenotype and incomplete penetrance may be due to factors such as ethnicity, environmental effects, and use of aminoglycosides. In addition, according to recent updates, mitochondrial transfer ribonucleic acid heteroplasmy variants might be responsible for this variation [8-11]. In Korean pedigrees, Hearing loss presented with wide penetrance and expressivity. The penetrance varied from 28.6% to 75%, (average of 60.8%) which were relatively higher than the Chinese population (29.5%) [7] but similar to Arab-Israeli and Spanish pedigrees (65.4% and 54.1% respectively) [12,13]. It has been reported that haplotype and functional variants are not related to the penetrance. It is believed that this condition places subject at risk for sensorineural hearing loss, but the severity (penetrance) of phenotype (hearing loss) is caused by multi-play of factors such as unidentified nuclear genes and aminoglycoside exposure [3].
In the present case report, for screening purposes, a recently developed genetic hearing disorder screening kit was used. The U-TOPвДҐHL Genotyping Kit, a real-time PCR-based method using the MeltingArray technique and PNA, was employed to screen 11 variants of 5 genes (GJB2, SLC26A4, MTRNR1, TMPRSS3, and CDH23). These genes covered varying degrees of sensorineural hearing loss and were selected for their high prevalence in Koreans which expected to cover up to 70% of prelingual genetic hearing loss cases in Koreans. The realtime PCR kit has 97.1% positive predictive value and 97.4 negative predictive value [1]. This test is covered by insurance (National Health Insurance Service of South Korea, in effect since July 2018) when the patientвАЩs condition meets one of the following standards; 1) confirmed congenital hearing loss, 2) pediatric hearing loss without middle ear pathology, 3) syndromic hearing loss, 4) confirmed inner ear anomaly in CT and MR, 5) progressive hearing loss with unknown origin, 6) hearing loss with familial history of genetic hearing loss.
The 1555A>G variant of the mitochondrial 12S rRNA gene (MTRNR1), the mutation in this case study, is found in 1.4% of the Korean hearing loss population. Of note, this mutation is the most common causative mutation of aminoglycosideinduced sensorineural hearing loss. We found that no antibiotics, including aminoglycosides, were taken by our patients. Because the origin of the mutation is the mitochondria, the hereditary pattern is maternal transmission (Fig. 1). The mother and son showed 1555A>G variant with homoplasmy by U-TOPвДҐHL Genotyping Kit and Sanger sequencing. According to Usami, et al., [14] approximately 3% of outpatients and 10% of cochlear implantation patients had the 1555A>G variant in Japan in 2000. The mechanism of pathogenesis is thought to be decreased efficiency of translation by 1555A>G, resulting in an impaired repair capability of the cochlea [15].
Meanwhile, there are other pathogenic variants in MTRNR1, such as 3243 A>G and 7445 A>G; however, they are rarer than 1555A>G [13]. Because the same mitochondrial variant shows a variable phenotype, as in our case family, functional study for pathogenesis encompassing a larger hearing loss family should be investigated. Estivill, et al. [12] mentioned that disease penetrance of 1555A>G is 50% in the 30s and 88% in 65-year-olds, showing age-dependent penetrance. Another explanation for variable expression is heteroplasmy, in which the proportion of normally functioning mitochondria affects the diverse degrees of hearing loss [16].
Although the U-TOPвДҐHL Genotyping Kit has flexibility for dealing with ethnicity-specific mutation spectra, it cannot detect novel variants. The novel variant could be a nondisease-causing single nucleotide polymorphism (SNP) or pathogenic SNP. More, larger-scale studies on normal controls using Sanger sequencing are needed to differentiate pathogenic SNPs.
There are many previous case reports and studies regarding this specific variant and hearing loss. Hearing loss in elderly without similar symptoms of oneвАЩs siblings as shown in a family of current case report is very common in otologic clinic and may not be a candidate for genetic hearing loss screenings. Detailed history taking, especially regarding family history of hearing loss, would be an important step. Nevertheless, due to the high price of the genetic workups, many hearing loss patients and their families who are suspected of having conditions of genetic origin cannot be thoroughly tested. But with this screening process at a reasonable price (in South Korea) patientвАЩs economic burden is much reduced. After the diagnosis, therapeutic plan for genetic hearing loss varies according to the characteristics of specific genetic conditions. In contrast to other genetic hearing losses which are not preventable, more detection of this genetic hearing losses that might be prevented by limiting aminoglycoside use at early ages might be possible and lead to a better quality of life in subjects. In addition, providing the information (by publication of this case report) about this specific genetic condition which could be aggravated by certain antibiotic use would help undetected patients with this condition.
ACKNOWLEDGMENTSThe English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/CbN7DY.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019R1F1A1058485).
NotesAuthor Contribution Conceptualization: Heejin Kim, Il-Seok Park. Data curation: Ju Eun Kim, Bo Ram Keum, Bin Kwon, Sung Min Park, Sung Won Li. Formal analysis: Ju Eun Kim, Heejin Kim. Methodology: Ju Eun Kim, Bo Ram Keum, Heejin Kim. Supervision: Heejin Kim, IlSeok Park. Visualization: Sung Won Li. WritingвАФoriginal draft: Ju Eun Kim. WritingвАФreview & editing: Sung Min Park, Heejin Kim, Il-Seok Park. Fig.¬†1.Pedigree of the family. This illustration is showing the pedigree of the family. Rectangles are male and circles are female. Filled circles and the subjects with hearing loss. The patient (mother with hearing loss) is depicted and marked as вАШPвАЩ. 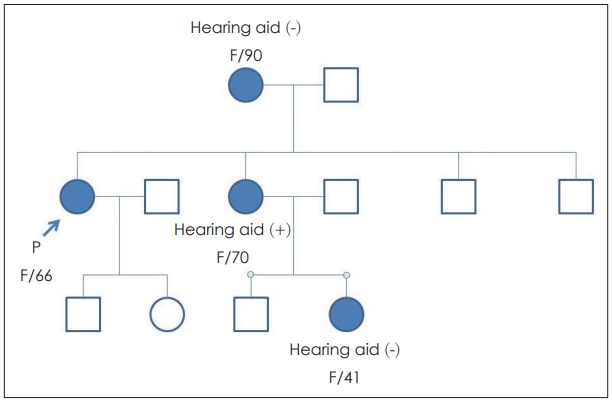
Fig. 2.Pure tone audiogram threshold of patient [X (blue line) is left ear; O (red line) is right ear]. (A) is pure tone audiometry of son showing normal hearing audiogram (B) is pure tone audiometry of mother showing both profound hearing loss (right worse). With left ear hearing aid (blue A), significant improvement of hearing is observed. 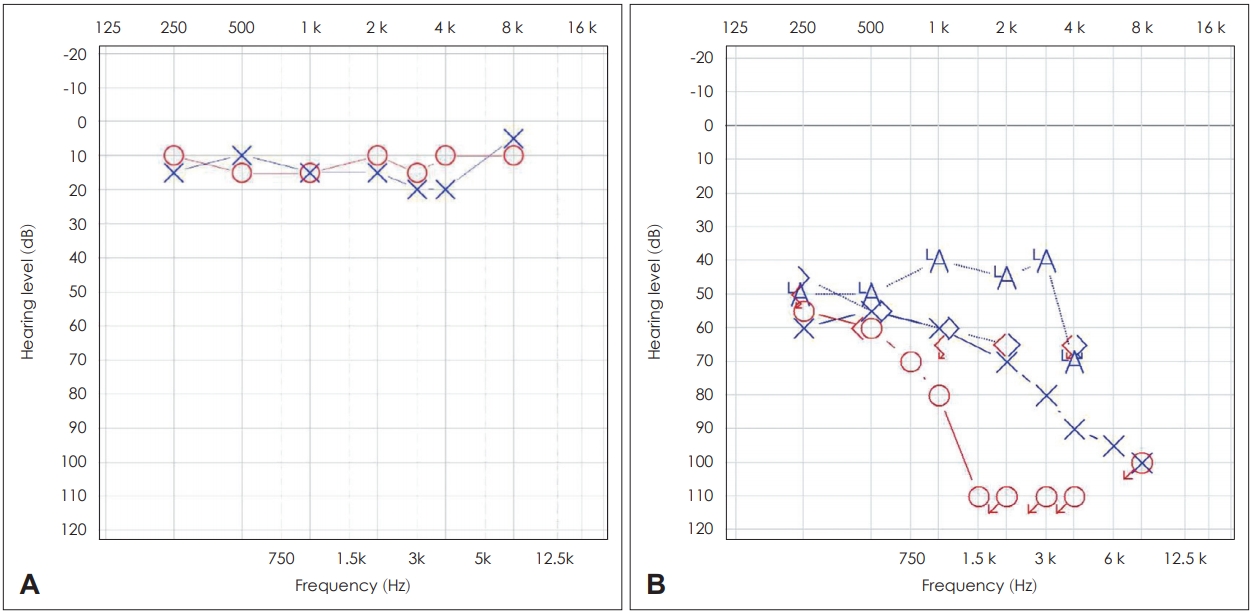
Table 1.The list of sequences of primers and PNA probes used in real-time PCR and primer sequences in Sanger sequencing REFERENCES1. Han KH, Kim AR, Kim MY, Ahn S, Oh SH, Song JH, et al. Establishment of a flexible real-time polymerase chain reactionbased platform for detecting prevalent deafness mutations associated with variable degree of sensorineural hearing loss in Koreans. PLoS One 2016;11(9):e0161756.
2. Bae JW, Kim DB, Choi JY, Park HJ, Lee JD, Hur DG, et al. Molecular and clinical characterization of the variable phenotype in Korean families with hearing loss associated with the mitochondrial A1555G mutation. PLoS One 2012;7(8):e42463.
3. Bai Y, Wang Z, Dai W, Li Q, Chen G, Cong N, et al. A six-generation Chinese family in haplogroup B4C1C exhibits high penetrance of 1555A>G-induced hearing loss. BMC Med Genet 2010;11:129.
4. Human H, Lombard D, de Jong G, Bardien S. A South African family with the mitochondrial A1555G mutation on haplogroup L0d. Biochem Biophys Res Commun 2009;382(2):390-4.
5. Dzhemileva LU, Posukh OL, Tazetdinov AM, Barashkov NA, Zhuravskiĭ SG, Ponidelko SN, et al. [Analysis of mitochondrial 12S rRNA and tRNA(Ser(UCN)) genes in patients with nonsyndromic sensorineural hearing loss from various regions of Russia]. Genetika 2009;45(7):982-91.
6. Lee MY, Ahn SY, Lee HJ, Jung JY, Rhee CK, Suh MW. Narrow band CE-Chirp auditory steady-state response is more reliable than the conventional ASSR in predicting the behavioral hearing threshold. Auris Nasus Larynx 2016;43(3):259-68.
7. Lu J, Qian Y, Li Z, Yang A, Zhu Y, Li R, et al. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A>G mutation. Mitochondrion 2010;10(1):69-81.
8. Zhu Y, Huang S, Kang D, Han M, Wang G, Yuan Y, et al. Analysis of the heteroplasmy level and transmitted features in hearing-loss pedigrees with mitochondrial 12S rRNA A1555G mutation. BMC Genet 2014;15:26.
9. Young WY, Zhao L, Qian Y, Li R, Chen J, Yuan H, et al. Variants in mitochondrial tRNAGlu, tRNAArg, and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three Han Chinese families with hearing loss. Am J Med Genet A 2006;140(20):2188-97.
10. Yuan H, Qian Y, Xu Y, Cao J, Bai L, Shen W, et al. Cosegregation of the G7444A mutation in the mitochondrial COI/tRNA(Ser(UCN)) genes with the 12S rRNA A1555G mutation in a Chinese family with aminoglycoside-induced and nonsyndromic hearing loss. Am J Med Genet A 2005;138A(2):133-40.
11. del Castillo FJ, Rodríguez-Ballesteros M, Martín Y, Arellano B, Gallo-Terán J, Morales-Angulo C, et al. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet 2003;40(8):632-6.
12. Estivill X, Govea N, Barceló E, Badenas C, Romero E, Moral L, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet 1998;62(1):27-35.
13. Bykhovskaya Y, Shohat M, Ehrenman K, Johnson D, Hamon M, Cantor RM, et al. Evidence for complex nuclear inheritance in a pedigree with nonsyndromic deafness due to a homoplasmic mitochondrial mutation. Am J Med Genet 1998;77(5):421-6.
14. Usami S, Abe S, Akita J, Namba A, Shinkawa H, Ishii M, et al. Prevalence of mitochondrial gene mutations among hearing impaired patients. J Med Genet 2000;37(1):38-40.
15. Usami S, Abe S, Kasai M, Shinkawa H, Moeller B, Kenyon JB, et al. Genetic and clinical features of sensorineural hearing loss associated with the 1555 mitochondrial mutation. Laryngoscope 1997;107(4):483-90.
16. Oh SH, Chang SO, Park HJ, Kim DY, Jeon SJ, Lim MJ, et al. Familial hearing loss associated with mtDNA A1555G mutation in Korea: 1 pedigree. Korean J Otolaryngol 1999;42(11):1353-8.
|
|
|||||||||||||||||||||||||||||||||||||||

 |
 |